SPARC Anatomy Working Group
Table of Contents
Overview
The Stimulating Peripheral Activity to Relieve Conditions (SPARC) effort is the result of National Institutes of Health’s (NIH) drive to map out the neural circuitry responsible for visceral control in higher vertebrates.
The management of multimodal anatomical knowledge is one of the mainstays of MAP-CORE’s effort in SPARC. At the core of this effort is the curation of computable and FAIR knowledge for the inferencing of multiscale parts and connectivity route pathways in support of SPARC metadata discovery.
The SPARC Anatomy Working Group (SAWG) is responsible for the integrity of anatomical knowledge in SPARC. In particular, it provides the relevant guidance and expertise about:
- Defining and naming anatomical terms
- Maintaining ontologies of anatomical knowledge
- Compiling and curating computable knowledge about multiscale routes pathways
This page provides access to the data generated by the SAWG which includes links to documentation about vocabulary services, term request and connectivity. This page will be continuously updated.
Terminology
Term lists
It is critical to the SPARC project that these terms lists be synched across the different contexts in which anatomical labels are applied. Towards that goal, we have gathered user requirements for annotation of data sets, images, flatmaps and models and created a set of term lists to make it easier for researchers to review and tools, e.g., MBF Biolucida, to use. Terms are extracted from existing ontologies whenever possible, primarily UBERON and FMA. UBERON, a cross species anatomical ontology, is the primary source of terms, but we also draw on FMA extensively for human terms as necessary. If the term does not exist in either ontology (UBERON or FMA) they are added to a tool built by UCSD for adding terms to ontologies called InterLex.
Below is a table containing all current anatomical term sets. They can be viewed as either a list in Google doc or a tree view in html. We posted them to Google Docs to make it easy for people to give feedback on individual terms by adding a comment. We also provide additional information on some of these term sets below.
| Annotation term set | Tabular | Tree | raw |
|---|---|---|---|
| Terms added to Interlex by SPARC | google sheet | trees/query | ttl |
| Brainstem | google sheet | trees/query | ttl |
| Spinal cord | google sheet | trees/query | ttl |
| Cranial nerves | google sheet | trees/query | ttl |
| Nerve roots of spinal cord segments | google sheet | trees/query | ttl |
| Nerves | google sheet | trees/query | ttl |
| Ganglia | google sheet | trees/query | ttl |
| Model and group terms | |||
| Atlas Nomenclature | google sheet | trees/query | ttl |
| ApiNATOMY | google sheet | trees/query | ttl |
| Peripheral Organ terms sets (MBF) | google sheet | trees/query | ttl |
| Minimal information model | google sheet | trees/query | ttl |
| Flatmap metadata | google sheet | trees/query | ttl |
| 3D scaffold | google sheet | trees/query | ttl |
Anatomical Terminology
Anatomy knowledge deals with the precise relationship between a human-readable anatomical term/label and the definition of that region (e.g. in the heart, community-wide consensus needs to be reached on the concept label/term “mitral valve” and the definition of what it is/what its boundaries are/over which range of organisms does homology of this structure hold). In addition, the identification (and, therefore, definition) of certain structures (e.g. subcellular) is dependent on a particular combination of preparing/staining/imaging/image processing techniques (i.e. a protocol). The navigation of such a wide combinatorial space of concepts, labels, definitions and protocols spawns a heterogeneity of approaches and, in consequence, opinions.
In this context, the SAWG provides community-supported leadership and arbitration on the accepted relationship between anatomical label and definition (and, by extension, the relationship between concepts - i.e. between a label-definition couplet and another).
In addition, part of this arbitration is to recommend and reconcile different topographical atlases that capture labelled regions within a coordinate system for some organ.
Another part of this arbitration is manifest via the term-request pipeline, in which a curator who has not found an appropriate label with which to annotate an image sub-region can submit a new term for global use within SPARC (and beyond).
- Documentation for adding terms to the SPARC vocabularies.
- SPARC has several methods for adding terms to the SPARC vocabularies, depending on the number of terms to be added. An overview of how to add to the SPARC vocabularies provides detailed instructions.
- For internal or advanced users who are familiar with the process, you can access the InterLex webpage here.
- SPARC vocabulary-building instructions
- Slide presentation outlining the strategy and procedures for building the SPARC vocabularies are
located here
- We keep a master list of all terms that have been added to Interlex by SPARC researchers for annotation of data, flatmaps or models. Each of these terms are reviewed by a knowledge curator to determine whether or not the term may already exist (for example, it might be a synonym of an existing term) or whether it should be formally added to the SPARC vocabularies. The SAWG, comprised of SPARC researchers and outside experts, provides advice to the knowledge curators about vocabulary additions.
- Slide presentation outlining the strategy and procedures for building the SPARC vocabularies are
located here
- Adoption of InterLex terminology services (for developers)
Up-to-date term lists for annotation in MBF tools by species and organ,including nomenclatures derived from particular atlases (together with acknowledgment/contact details for leads responsible for each list);
-see table above for CNS and peripheral nervous system structures.
- Up-to-date listing of terms applied to SPARC1 metadata annotation
- Up-to-date listing of terms applied to flatmap metadata annotation
- Up-to-date listing of terms applied to ApiNATOMY metadata annotation google sheet
- Up-to-date listing of terms applied to 3D scaffold metadata annotation
- Coming soon…
- Searchable full reference list of terms (drawn from UBERON/FMA/Nomina/atlases)
- Coming soon…
Connectivity
The development of multiscale route knowledge graphs is leveraging the ApiNATOMY knowledge representation. A graphical depiction of an ApiNATOMY model relevant to defensive breathing and Superior Cervical Ganglion physiology is shown below.
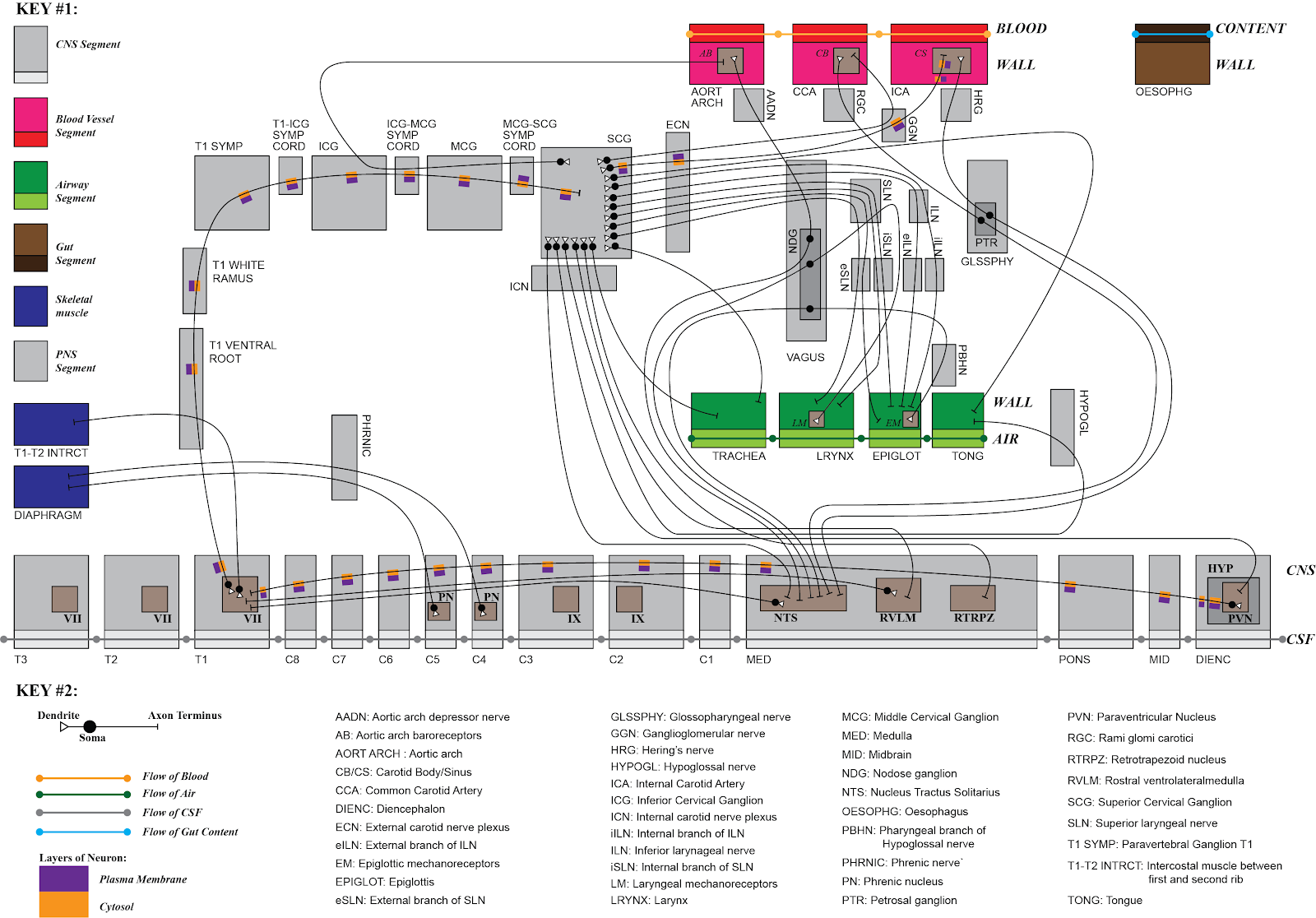
ApiNATOMY is used to build routing and connectivity graphs for anatomical entities. Such graphs support queries that, for instance, identify neural connections that course through a tract, nerve or ganglion. ApiNATOMY-based knowledge, therefore, allows the SPARC user to determine the nuclei/grey matter regions affected by the transection of a nerve or the stimulation of a ganglion. In addition, the same routing information leveraged by the flatmap GUI may be used to discover metadata to SPARC experimental data or simulation models that locate along the route of a tract, nerve or ganglion.
- Catalogue of ApiNATOMY connectivity models
- Coming soon
- Text-based querying of ApiNATOMY models (and SPARC1 metadata discovery)
- Flatmap-based querying of ApiNATOMY models (and SPARC1 metadata discovery)
- Coming soon
Expertise
- Diagram/bibliography repository for flatmap construction
- TBA
- SAWG outside expert working group: Current members
- Jackie Bresnahan, Ph.D.
Professor, Neurological Surgery, University of California, San Francisco
Dr. Bresnahan's research focuses on understanding the biological underpinnings of neurotrauma, particularly spinal cord injury, with the goal of improving recovery for individuals who suffer damage to the nervous system. Her laboratory has developed a number of models to study cellular systems and behavioral changes that occur as a consequence of injury. - Gary Mawe, Ph.D.
Samuel W. Thayer Professor of Neurological Sciences, University of Vermont
Dr. Mawe's research focuses on the neurobiology of GI function in health and disease states and uses a variety of experimental approaches including electrophysiology, calcium imaging, immunocytochemistry and molecular biology. He is actively involved in teaching anatomy, physiology and comparative neurobiology to undergraduates, graduates students, medical students, and residents.
- Jackie Bresnahan, Ph.D.
- Discussion forum for requirements collection, gap analysis, feedback
- Coming soon
- Documents section: reports and other publications
- TBA
- Events and other announcements
- TBA
- Challenges: e.g. coursing of visceral autonomic nerves
- TBA